A successful ChIP-seq experiment requires an antibody that recognizes the correct target protein in all sequence contexts across the entire genome. Good antibody performance in ChIP-qPCR does not necessarily mean the antibody will perform well for ChIP-seq, because ChIP-seq requires more extensive capture of the target protein across a large number of gene loci. To provide antibodies that are proven effective for ChIP-seq, CST validates recombinant rabbit monoclonal antibodies for ChIP-seq using the SimpleChIP® Enzymatic and SimpleChIP® Sonication Chromatin IP protocols followed by next-generation sequencing (NGS).
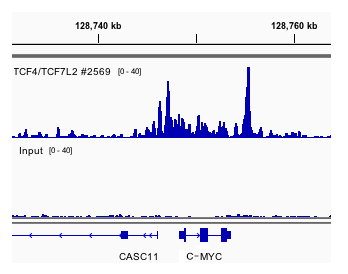
Chromatin immunoprecipitations were performed with cross-linked chromatin from HCT116 cells and TCF4/TCF7L2 (C48H11) Rabbit mAb #2569, using SimpleChIP® Enzymatic Plus Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figure shows a specific binding of TCF4/TCF7L2 across c-MYC, a known target gene of TCF4/TCF7L2 (upper track), compared to Input control (lower track). The average signal:noise ratio across the c-MYC gene is 21.58.
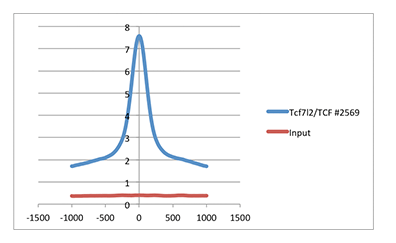
Chromatin immunoprecipitations were performed with cross-linked chromatin from HCT116 cells and TCF4/TCF7L2 (C48H11) Rabbit mAb #2569, using SimpleChIP® Enzymatic Plus Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. As shown in the figure, the average signal:noise ratio across the whole genome for this experiment, comparing the TCF4/TCF7L2 enrichment (blue line) to the Input control (red line), is 4.44.
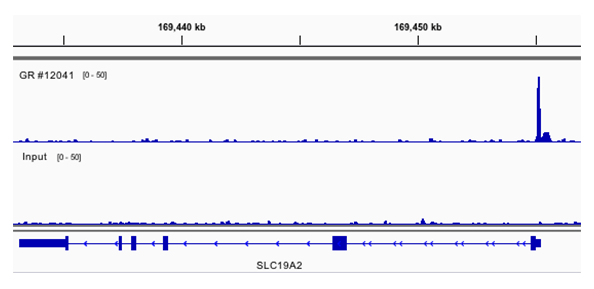
Chromatin immunoprecipitations were performed with cross-linked chromatin from A549 cells cultured in media with 5% charcoal-stripped FBS for 3 d and then treated with 100 nM dexamethasone for 1 hour and Glucocorticoid Receptor (D6H2L) XP® Rabbit mAb #12041, using SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. The figure shows the specific binding of GR across SLC19A2, a known target gene of GR (upper track), compared to the input control (lower track). Forty-one percent (41%) of the GR antibody-enriched DNA fragments contain the known glucocorticoid response element binding motif GNACANNNTGTNC.
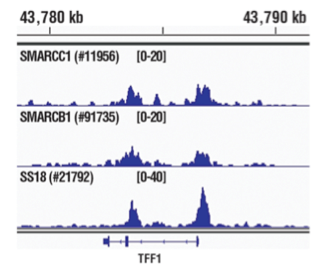
Chromatin immunoprecipitations were performed with cross-linked chromatin from MCF7 cells grown in phenol red free medium and 5% charcoal stripped FBS for 4 d, followed by treatment with β-estradiol (10 nM, 45 min) and either SMARCC1/BAF155 (D7F8S) Rabbit mAb #11956, SMARCB1/BAF47 (D8M1X) Rabbit mAb #91735, or SS18 (D6I4Z) Rabbit mAb #21792, using SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. SMARCC1/BAF155, SMARCB1/BAF47, and SS18 are all subunits of SWI/SNF complex. As expected, all three protein subunits of the SWI/SNF complex show overlapping binding across pS2/TFF1, a known target gene of the SWI/SNF complex.